载入数据
1Sys.setlocale('LC_ALL','C') 2load(file = "F:/Bioinfor_project/Breast/AS_research/AS/result/hubgene.Rdata") 3head(data) 4require(cowplot) 5require(tidyverse) 6require(ggplot2) 7require(ggsci) 8require(ggpubr) 9mydata<-data %>% 10## 基因表达数据gather,gather的范围应调整11gather(key="gene",value="https://www.itzhengshu.com/wps/Expression",CCL14:TUBB3) %>% 12##13dplyr::select(ID,gene,Expression,everything()) 14head(mydata)## 每个基因作为一个变量的宽数据创建带有pvalue的箱线图
- 参考资料
- 展示绘图细节控制
1p <- ggboxplot(mydata, x = "group", y = "Expression",2color = "group", palette = "jama",3add = "jitter")4#Add p-value5pstat_compare_means()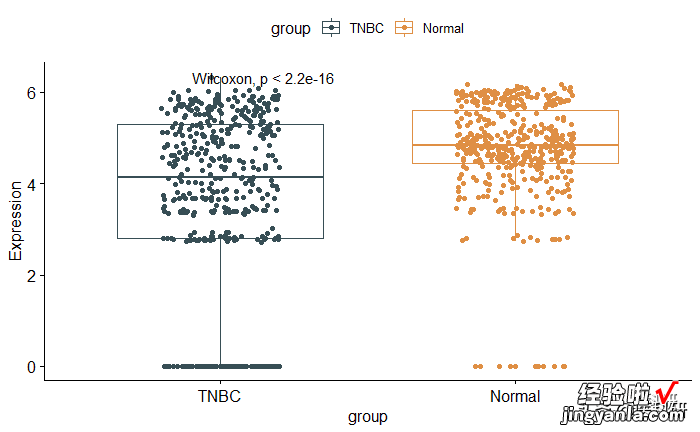
【R语言ggboxplot-一文掌握箱线图绘制所有细节】image.png
改变统计方法
1# Change method2pstat_compare_means(method = "t.test")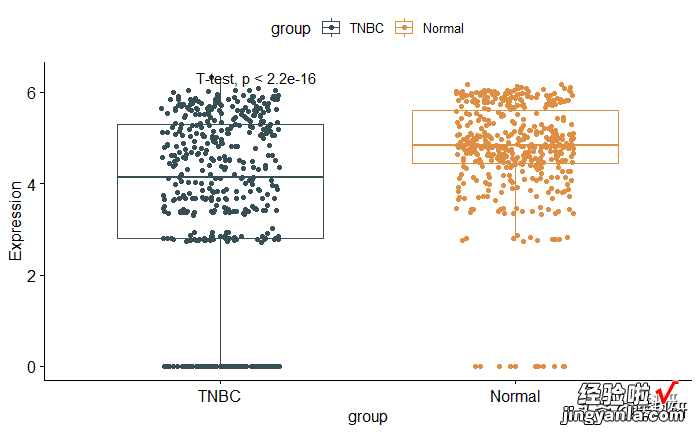
image.png
统计学意义标注
- label="p.signif"
- p.format等
- label.x标注位置
1pstat_compare_means( label = "p.signif")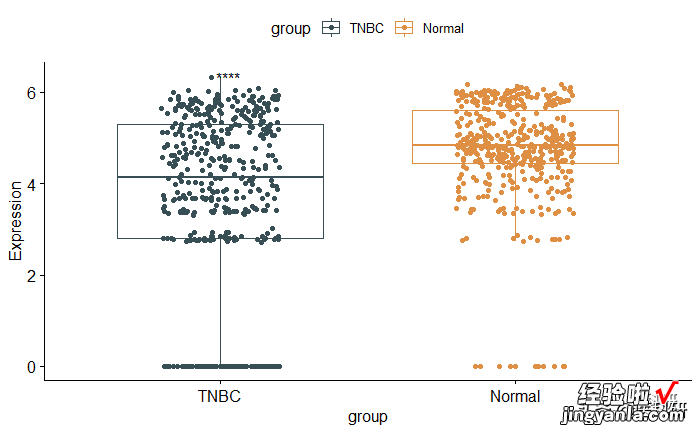
image.png
多组比较
- 给出global pvalue
1# Default method = "kruskal.test" for multiple groups2ggboxplot(mydata, x = "gene", y = "Expression",3color = "gene",add="jitter", palette = "jama") 4stat_compare_means()56# Change method to anova7ggboxplot(mydata, x = "gene", y = "Expression",8color = "gene", add="jitter", palette = "jama") 9stat_compare_means(method = "anova")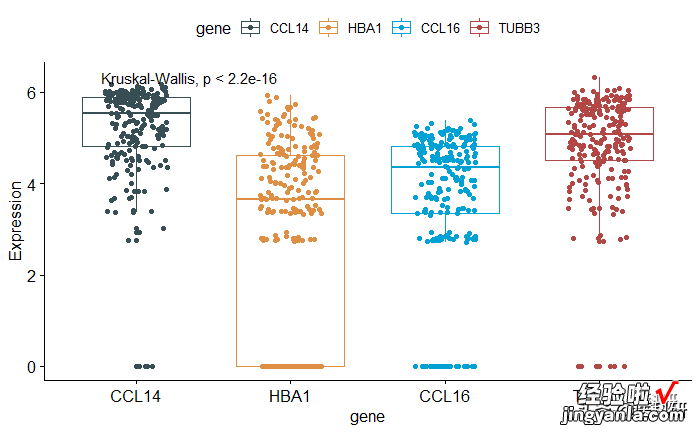
image.png
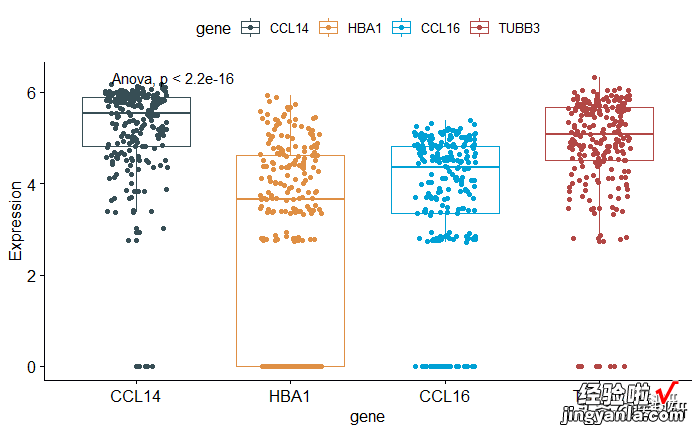
image.png
指定比较
- 配对比较:会完成各个变量的比较,默认wilcox.test法,可修改
- my_comparisions:可以指定自己想要进行的比较
- 指定参考组,进行比较
1require(ggpubr) 2compare_means(Expression ~ gene,data = mydata) 3 4## 指定自己想要的比较 5# Visualize: Specify the comparisons you want 6my_comparisons <- list( c("CCL14", "HBA1"), c("HBA1", "CCL16"), c("CCL16", "TUBB3") ) 7ggboxplot(mydata, x = "gene", y = "Expression", 8color = "group",add = "jitter", palette = "jama")9stat_compare_means(comparisons = my_comparisons)## Add pairwise comparisons p-value10#stat_compare_means()# Add global p-value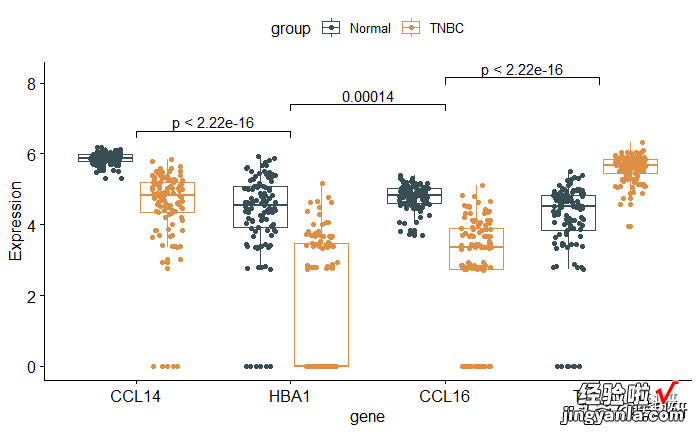
image.png
指定参考组
指定CCL14作为参考组与其它各组比较
ref.group
1compare_means(Expression ~ gene,data = https://www.itzhengshu.com/wps/mydata, ref.group ="CCL14", 2method = "t.test") 3# Visualize 4mydata %>_ilter(group=="TNBC") %>% # 筛选TNBC数据 6ggboxplot( x = "gene", y = "Expression", 7color = "gene",add = "jitter", palette = "nejm")8stat_compare_means(method = "anova")# Add global p-value 9stat_compare_means(label = "p.signif", method = "t.test",10ref.group = "CCL14")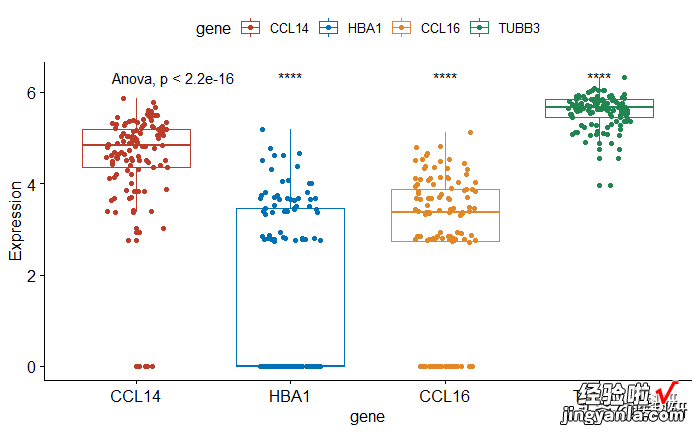
image.png
多基因分面
按另外一个变量分组比较
1## 比较各个基因在TNBC与Normal表达 2compare_means( Expression ~ group, data = https://www.itzhengshu.com/wps/mydata,3group.by ="gene") 4# Box plot facetted by "gene" 5p <- ggboxplot(mydata, x = "group", y = "Expression", 6color = "group", palette = "jco", 7add = "jitter", 8facet.by = "gene", short.panel.labs = FALSE) 9# Use only p.format as label. Remove method name.10pstat_compare_means(label = "p.format")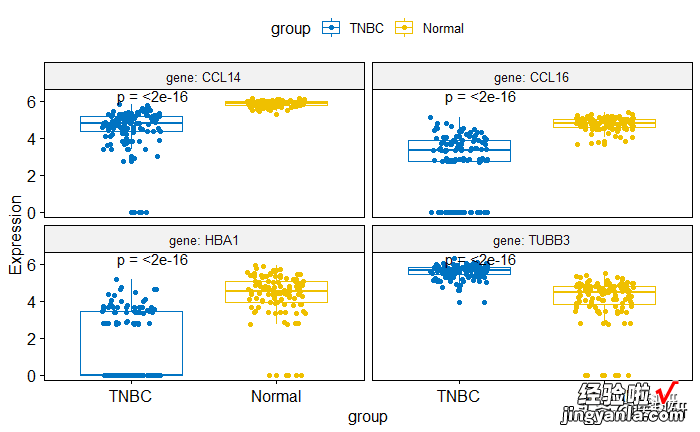
image.png
将pvalue换成星号
- hide.ns = TRUE.参数可隐藏ns
1pstat_compare_means(label ="p.signif", label.x = 1.5)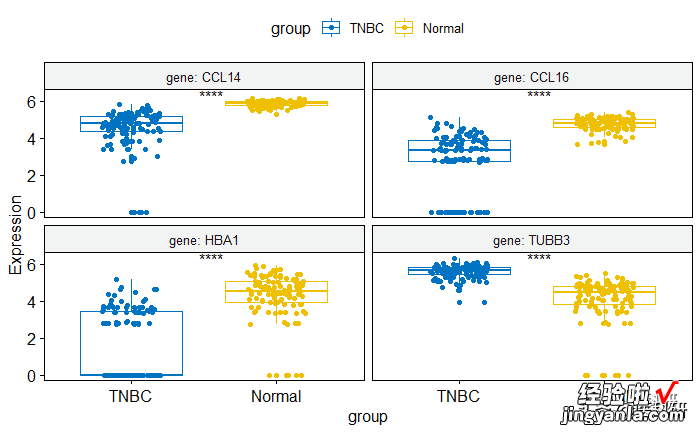
image.png
将各个图绘制在一张图中
1p <- ggboxplot(mydata, x = "gene", y = "Expression",2color = "group", palette = "nejm",3add = "jitter")4pstat_compare_means(aes(group = group))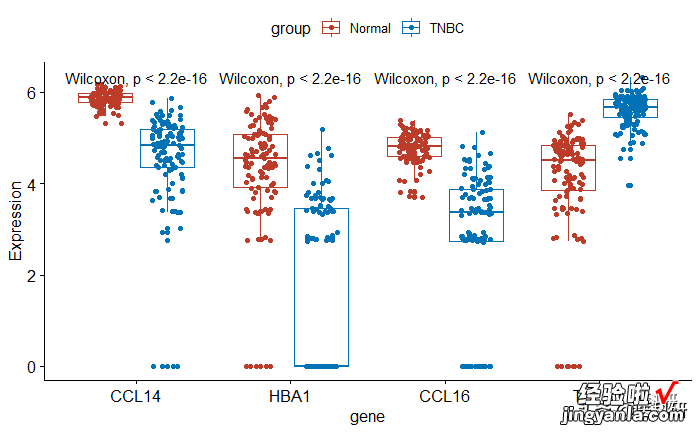
image.png
修改下pvalue展示的方式
1# Show only p-value2pstat_compare_means(aes(group = group), label = "p.format")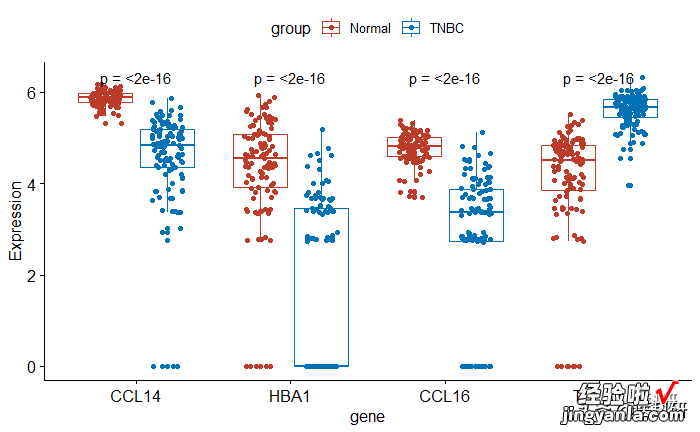
image.png
用星号表示pvalue
1# Use significance symbol as label2pstat_compare_means(aes(group = group), label = "p.signif")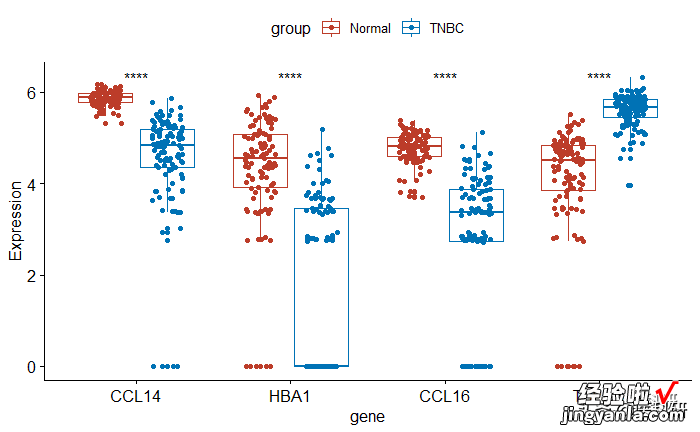
image.png
配对样本比较
要求x,y具有相同的样本数,进行一一配对比较
1head(ToothGrowth) 2compare_means(len ~ supp, data = https://www.itzhengshu.com/wps/ToothGrowth,3group.by ="dose", paired = TRUE) 4# Box plot facetted by "dose" 5p <- ggpaired(ToothGrowth, x = "supp", y = "len", 6color = "supp", palette = "jama",7line.color = "gray", line.size = 0.4, 8facet.by = "dose", short.panel.labs = FALSE) 9# Use only p.format as label. Remove method name.10pstat_compare_means(label = "p.format", paired = TRUE)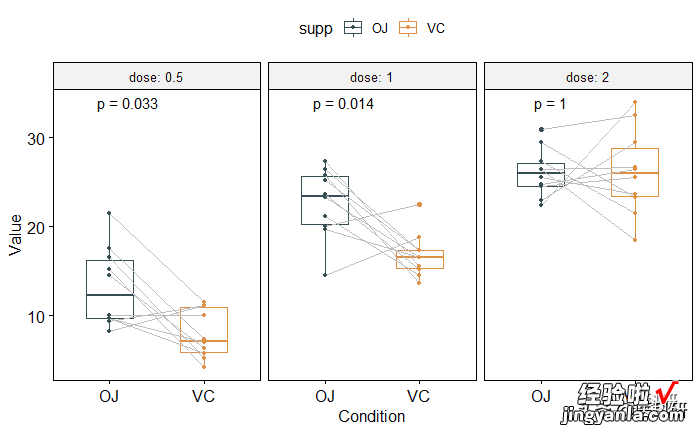
image.png
封装为函数命名为group_box
- 功能:已经选定的基因绘制箱线图
- - 参数1:group分组变量,可以是自己所有感兴趣的变量
- 参数2:mydata为整理好的清洁数据,gene为长数据(gather版本)
1head(mydata) 2group_box<-function(group=group,data=https://www.itzhengshu.com/wps/mydata){ 3p <- ggboxplot(mydata, x ="gene", y = "Expression", 4color = group,5palette = "nejm", 6add = "jitter") 7pstat_compare_means(aes(group = group)) 8} 910## 11group_box(group="PAM50",data = https://www.itzhengshu.com/wps/mydata)封装为函数命名为group_box
- 功能:已经选定的基因绘制箱线图
- - 参数1:group分组变量,可以是自己所有感兴趣的变量
- 参数2:mydata为整理好的清洁数据,gene为长数据(gather版本)
1head(mydata) 2group_box<-function(group=group,data=https://www.itzhengshu.com/wps/mydata){ 3p <- ggboxplot(mydata, x ="gene", y = "Expression", 4color = group,5palette = "nejm", 6add = "jitter") 7pstat_compare_means(aes(group = group)) 8} 910## 11group_box(group="PAM50",data = https://www.itzhengshu.com/wps/mydata)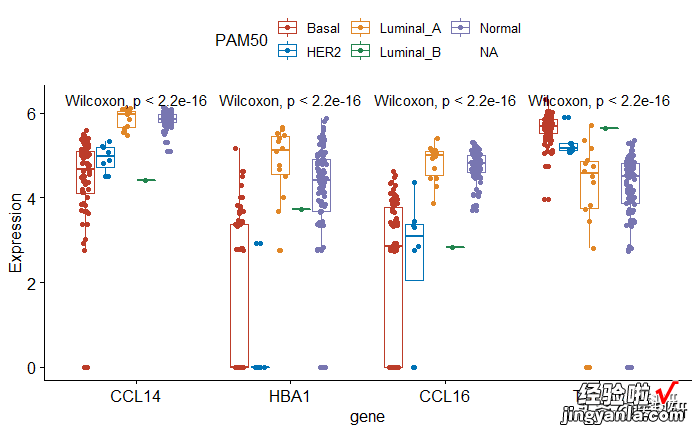
image.png
封装函数gene_box
- 目的功能:对感兴趣的基因绘制和分组绘制boxplot
- 注意这时使用的应该是基因的宽数据,因为涉及到单个基因作为变量
1head(data) 2usedata<-data 3## 封装函数 4gene_box<-function(gene="CCL14",group="group",data=https://www.itzhengshu.com/wps/usedata){ 5p <- ggboxplot(data, x = group, y = gene, 6ylab = sprintf("Expression of %s",gene), 7xlab = group, 8color = group,9palette = "nejm",10add = "jitter")11pstat_compare_means(aes(group = group))12}1gene_box(gene="CCL14")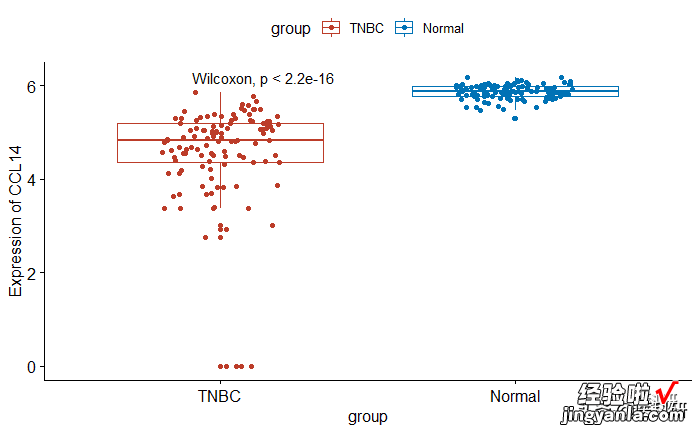
image.png
牛刀小试
1gene_box(gene="CCL16",group="PAM50")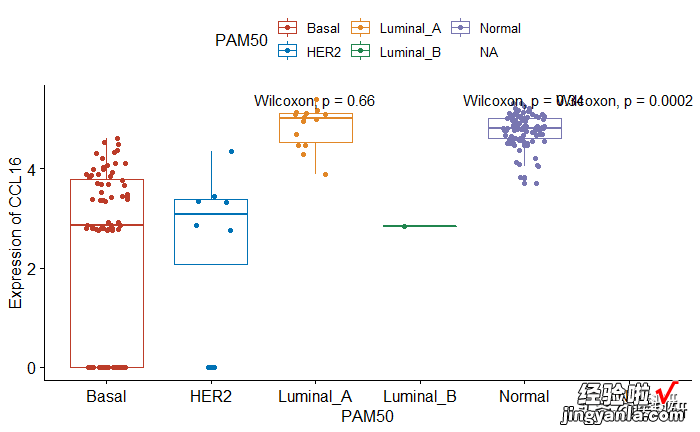
image.png
批量绘制
- 目的功能:绘制任意基因,任意分组,批量绘制一气呵成了
- 封装函数 lapply批量绘制无敌
- 在lapply中的函数参数设置,不在原函数中,而是直接放置在lapply中
- do.call中参数1为函数,c()包含原函数的参数设置,同样参数设置不在原函数中
1require(gridExtra)2head(data)34## 需要批量绘制的基因名5name<-colnames(data)[3:6]6## 批量绘图7p<-lapply(name,gene_box,group = "T_stage")8## 组图9do.call(grid.arrange,c(p,ncol=2))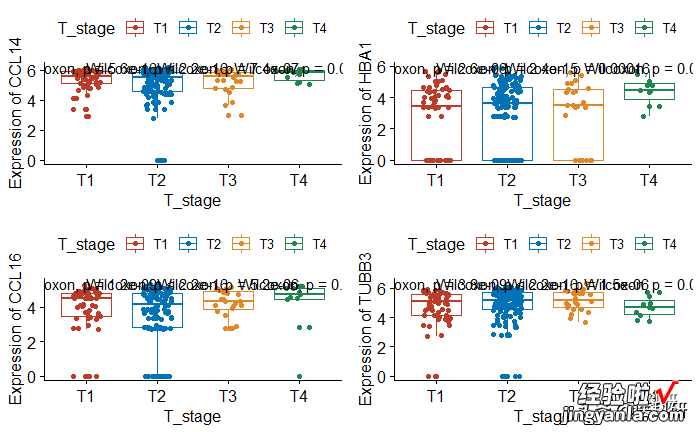
image.png